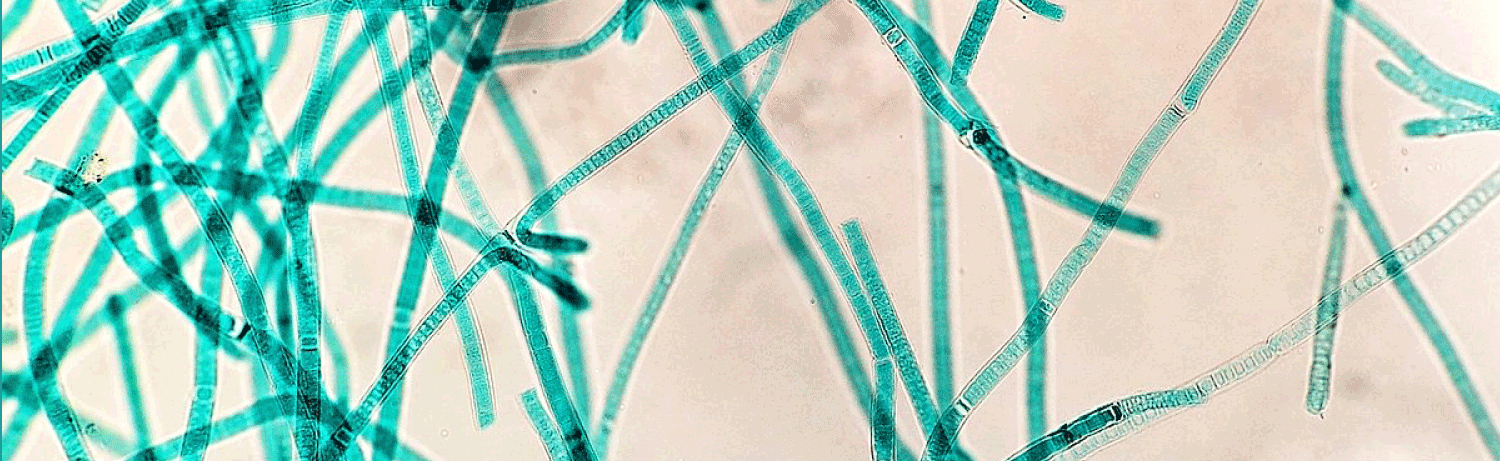
Another important milestone for the project has been published in the journal Nature Catalysis, in a paper that describes one of the promising pathways developed in FutureAgriculture to bypass the photorespiration.
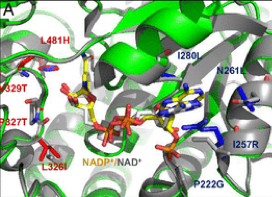
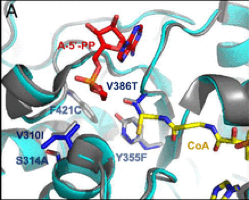
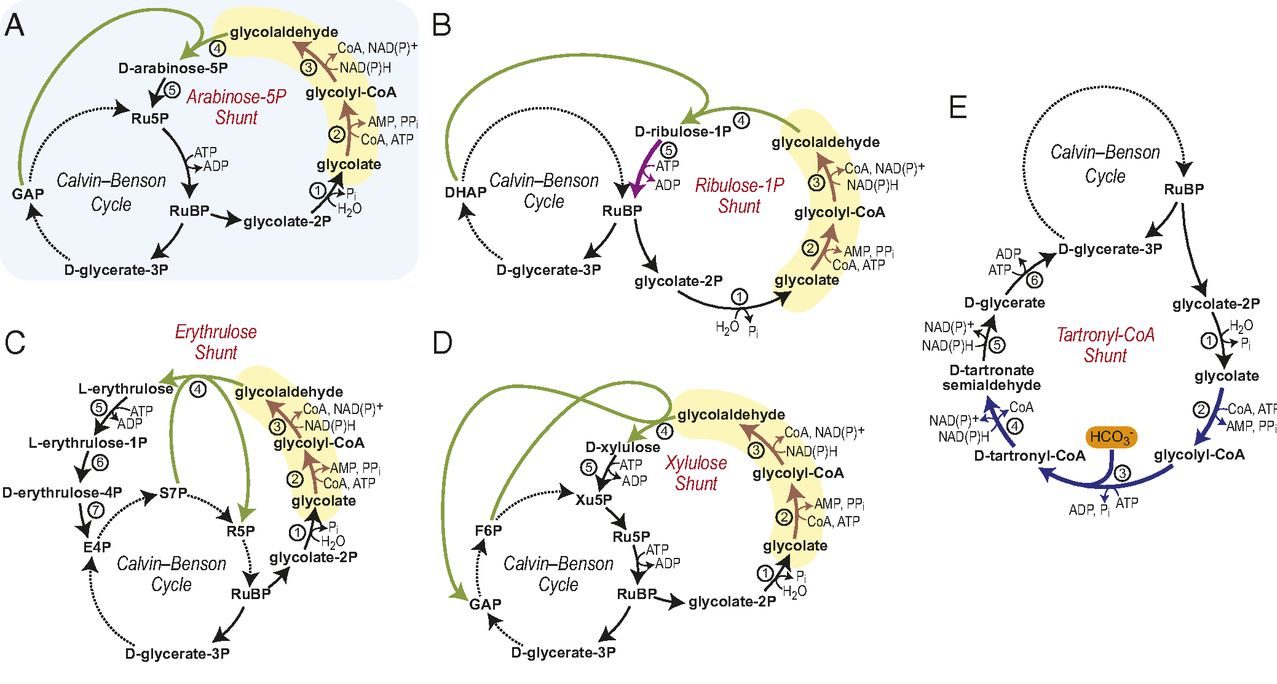
The team led by Tobias Erb from the Max Planck Institute for Terrestrial Microbiology has developed a synthetic photorespiratory bypass that represents an alternative to natural photorespiration. Together with the Consortium, the Erb’s lab has designed the so-called tartronyl-CoA (TaCo) pathway that is much shorter than natural photorespiration and requires only 5 instead of 11 enzymes. The perhaps greatest benefit of the TaCo pathway is that it fixes CO2 instead of releasing it, as it happens in natural photorespiration. As a result, the TaCo pathway is more energy-efficient than any other proposed photorespiratory bypass to date.
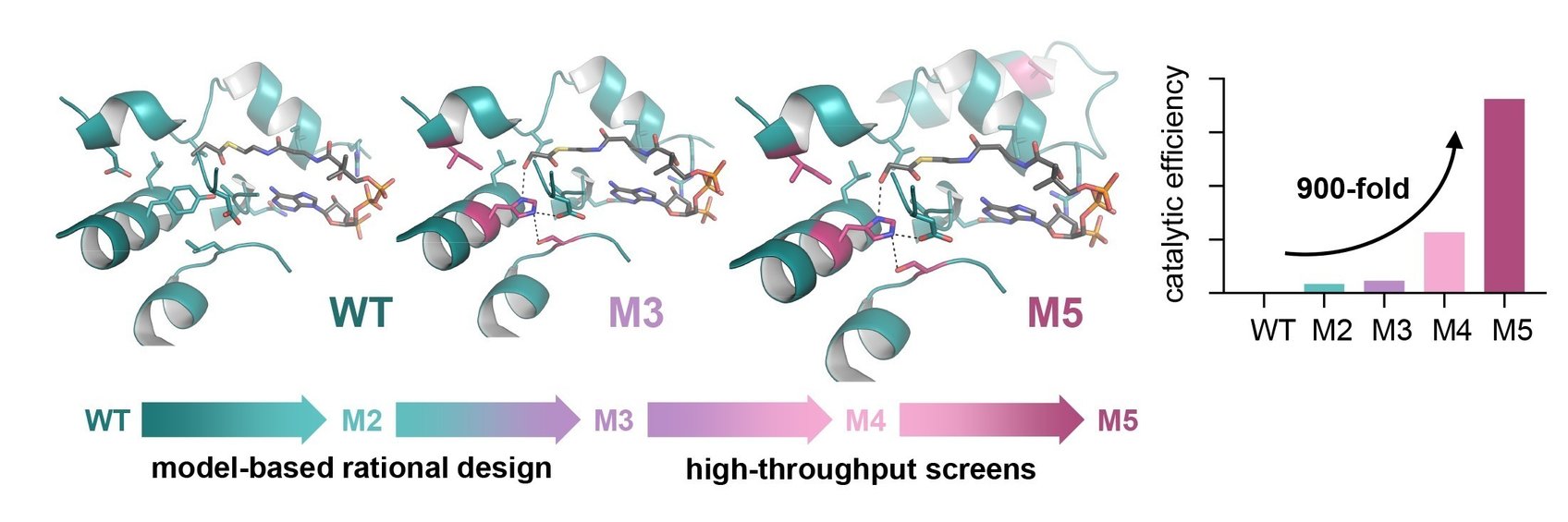
Building the TaCo pathway was a scientific journey that has led the researchers from the computational model through enzymatic engineering, microfluidic high-throughut screening, cryo-EM-technology towards the successful in vitro implementation of a new-to-nature metabolic connection that opens up new possibilities for CO2 fixation and the production of value-added compounds. “The main challenge in realizing the TaCo pathway was to find all the required enzymes,” Marieke Scheffen, Postdoctoral researcher in Tobias Erb`s group and lead author of the study, recalls. “It meant that we had to look for enzymes that perform similar reactions and then “teach” them to perform the desired reaction.”
For more information please check the original news by MPI and the scientific paper.
Reference:








 What’s the story behind our project? Read on to discover it in the words of our project coordinator, Arren Bar-Even from MPI-MP. He wrote an article for Science Trends providing his overview of the project, its objectives and the legacy that he hopes to leave. The full article can be found
What’s the story behind our project? Read on to discover it in the words of our project coordinator, Arren Bar-Even from MPI-MP. He wrote an article for Science Trends providing his overview of the project, its objectives and the legacy that he hopes to leave. The full article can be found 

 Our partner, Tobias Erb from the Max Planck Institute for Terrestrial Microbiology have been awarded the prestigious
Our partner, Tobias Erb from the Max Planck Institute for Terrestrial Microbiology have been awarded the prestigious 
 We are currently collaborating with MedioMix to create the official video for FutureAgriculture! The first shooting day was at the MPI-MP in Potsdam – it has been a long day among laboratories, greenhouses and finding the right location for interviews. The official video for FutureAgriculture will be released soon, in the meantime, you can check our other videos on the dedicated
We are currently collaborating with MedioMix to create the official video for FutureAgriculture! The first shooting day was at the MPI-MP in Potsdam – it has been a long day among laboratories, greenhouses and finding the right location for interviews. The official video for FutureAgriculture will be released soon, in the meantime, you can check our other videos on the dedicated  Unleashing the technical present / Nov 30 – Dec 02. Researchers & artists met for this experimental conference in order to explore the meaning and implications of the technosphere. Arren Bar-Even was invited to the SEEDS session – watch the full performance
Unleashing the technical present / Nov 30 – Dec 02. Researchers & artists met for this experimental conference in order to explore the meaning and implications of the technosphere. Arren Bar-Even was invited to the SEEDS session – watch the full performance  Our project coordinator got the chance to interact with artists and scientists on the topic “ESTHETICS get SYNTHETIC: Knowledge Link Through Art & Science” during the workshop organized by KLAS at the
Our project coordinator got the chance to interact with artists and scientists on the topic “ESTHETICS get SYNTHETIC: Knowledge Link Through Art & Science” during the workshop organized by KLAS at the  One of our partners, the Max Plack Institute for Molecular Plant Physiology – MPI-MP, has been selected among the 4 finalists for the European Innovation Radar prize 2017 under the category Excellent Science. This category selects the best cutting-edge science underpinning tomorrow's technological advances. Thanks to this initiative, the FutureAgriculture's team got the chance to pitch their plans for going to market with their EU-funded tech to a jury of experts at the ICT Proposers' Day in Budapest (9 November 2017). More info
One of our partners, the Max Plack Institute for Molecular Plant Physiology – MPI-MP, has been selected among the 4 finalists for the European Innovation Radar prize 2017 under the category Excellent Science. This category selects the best cutting-edge science underpinning tomorrow's technological advances. Thanks to this initiative, the FutureAgriculture's team got the chance to pitch their plans for going to market with their EU-funded tech to a jury of experts at the ICT Proposers' Day in Budapest (9 November 2017). More info