The National Academy of Sciences in Germany have selected artificial photosynthesis as one of the key technologies to keep an eye on. Tobias Erb was part of the working group that aimed at evaluating the state-of-the-art of this technology and its future perspectives. We asked him to summarize this experience for us.
The National Academy of Sciences in Germany have selected artificial photosynthesis as one of the key technologies to keep an eye on. Tobias Erb was part of the working group that aimed at evaluating the state-of-the-art of this technology and its future perspectives. We asked him to summarize this experience for us.
T: Over the last two years, I was part of the group of experts, coming from different disciples (biology, physics, chemistry, material sciences, etc), that analysed the state of the art of the artificial photosynthesis for the German Academy of Sciences. The field is actually vast, it includes projects like Future Agriculture, in which synthetic biology is used to improve the natural photosynthesis, and others that try to mimic the basic principles of photosynthesis by combining chemical catalysts with photovoltaic elements.
We have been asked to imagine what we could achieve with this technology in the next ten, twenty, and thirty years, respectively, and what it will require to move the field forward. We all agreed that synthetic biology is an important technology in developing synthetic photosynthesis. There is an innovative and disruptive potential in synthetic biology and thus it is important to keep the vision up and going. Some projects are very risky, like FA, as there is no sure outcome. However, you need to push the limits of technologies and see what it is achievable – that’s why national and Europe projects are fundamental to keep the disciple up and running.
What is so disruptive about synthetic biology now?
T: In Synthetic Biology, we build something new by recombining biological parts. We can implement very refined and rational changes to build something that Nature has not invented yet, following our intuition. The evolution of the discipline doesn’t differ from what happened in other fields too. Chemistry, for example, started by studying the building blocks of matter i.e. elements, atoms etc. Then it moved forward to synthetize new compounds using the same building blocks. And that’s how we ended up with modern textiles, colours, drugs – and an entire range of new materials that are non-natural but commonly used.
That’s exactly what it is happening now in biology: we started by studying the building blocks of the biological material: enzyme, cells, DNA, etc. Now we can use them to synthetize something new with the additional advantage that biological materials are self-optimizing and environmentally sustainable compared to chemistry.
What the future holds for synthetic biology?
T: The market potential for synthetic biology is estimated at around 38 billion euros for 2020; it will be a key technology in the near future and there is no way to skip this. Nowadays you can look up gene sequences in public databases, use modeling software to make predictions how this gene will perform in the context of a cell and you can order the gene for little more than 100 euros online. Prices will probably drop even more and it’s going to be easier and cheaper to use this technology.
At this point we are witnessing the merge between information technology and biology – as scientists, we aim at developing future technologies but citizens and policymakers are the ones that will decide if and how these technologies should be used. This is still an ongoing process because we need to make sure that the technology is feasible, safe and accepted by people. For that, we need to openly communicate throughout the entire process. It’s important that citizens realize that they are part of the decision process and that scientists are listening to the needs and fears of people. Facts and evidence work well in science but to create a fruitful discussion between science and society the best strategy is listening.











 Patrik Jones is the group leader of the Microbial Metabolic Engineering lab at the Imperial College London. His lab aims at engineering metabolic solution to convert CO2 into biomass and other sustainable chemicals that can replace less sustainable products. We asked him to share his point of view on sustainability.
Patrik Jones is the group leader of the Microbial Metabolic Engineering lab at the Imperial College London. His lab aims at engineering metabolic solution to convert CO2 into biomass and other sustainable chemicals that can replace less sustainable products. We asked him to share his point of view on sustainability.

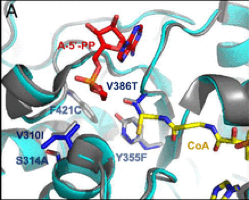
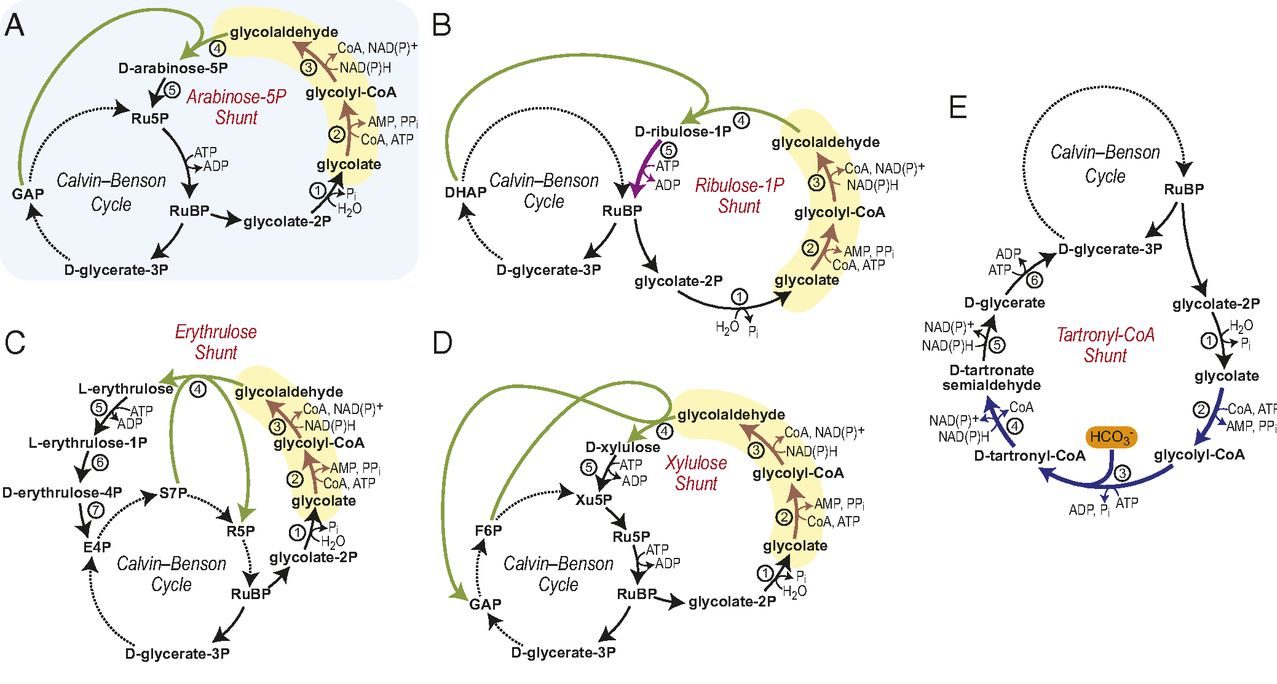
 What’s the story behind our project? Read on to discover it in the words of our project coordinator, Arren Bar-Even from MPI-MP. He wrote an article for Science Trends providing his overview of the project, its objectives and the legacy that he hopes to leave. The full article can be found
What’s the story behind our project? Read on to discover it in the words of our project coordinator, Arren Bar-Even from MPI-MP. He wrote an article for Science Trends providing his overview of the project, its objectives and the legacy that he hopes to leave. The full article can be found 









 The National Academy of Sciences in Germany have selected artificial photosynthesis as one of the key technologies to keep an eye on. Tobias Erb was part of the working group that aimed at evaluating the state-of-the-art of this technology and its future perspectives. We asked him to summarize this experience for us.
The National Academy of Sciences in Germany have selected artificial photosynthesis as one of the key technologies to keep an eye on. Tobias Erb was part of the working group that aimed at evaluating the state-of-the-art of this technology and its future perspectives. We asked him to summarize this experience for us.